The goal of this book is to help you learn about JBoss DNA and how you can use it in your own applications to get the most out of your JCR repositories.
The first part of the book starts out with an introduction to content repositories and an overview of the JCR API, both of which are important aspects of JBoss DNA. This is followed by an overview of the JBoss DNA project, its architecture, and a basic roadmap for what's coming next.
The next part of the book covers how to download and build the examples, how to use JBoss DNA with existing repositories, and how to build and use custom sequencers.
If you have any questions or comments, please feel free to contact JBoss DNA's user mailing list or use the user forums . If you'd like to get involved on the project, join the mailing lists , download the code and get it building, and visit our JIRA issue management system . If there's something in particular you're interested in, talk with the community - there may be others interested in the same thing.
There are a lot of ways for applications to store information persistently so that it can be accessed at a later time and by other processes. The challenge developers face is how to use an approach that most closely matches the needs of their application. This choice becomes more important as developers choose to focus their efforts on application-specific logic, delegating much of the responsibilities for persistence to libraries and frameworks.
Perhaps one of the easiest techniques is to simply store information in files . The Java language makes working with files relatively easy, but Java really doesn't provide many bells and whistles. So using files is an easy choice when the information is either not complicated (for example property files), or when users may need to read or change the information outside of the application (for example log files or configuration files). But using files to persist information becomes more difficult as the information becomes more complex, as the volume of it increases, or if it needs to be accessed by multiple processes. For these situations, other techniques often have more benefits.
Another technique built into the Java language is Java serialization , which is capable of persisting the state of an object graph so that it can be read back in at a later time. However, Java serialization can quickly become tricky if the classes are changed, and so it's beneficial usually when the information is persisted for a very short period of time. For example, serialization is sometimes used to send an object graph from one process to another. Using serialization for longer-term storage of information is more risky.
One of the more popular and widely-used persistence technologies is the relational database. Relational database management systems have been around for decades and are very capable. The Java Database Connectivity (JDBC) API provides a standard interface for connecting to and interacting with relational databases. However, it is a low-level API that requires a lot of code to use correctly, and it still doesn't abstract away the DBMS-specific SQL grammar. Also, working with relational data in an object-oriented language can feel somewhat unnatural, so many developers map this data to classes that fit much more cleanly into their application. The problem is that manually creating this mapping layer requires a lot of repetitive and non-trivial JDBC code.
Object-relational mapping libraries automate the creation of this mapping layer and result in far less code that is much more maintainable with performance that is often as good as (if not better than) handwritten JDBC code. The new Java Persistence API (JPA) provide a standard mechanism for defining the mappings (through annotations) and working with these entity objects. Several commercial and open-source libraries implement JPA, and some even offer additional capabilities and features that go beyond JPA. For example, Hibernate is one of the most feature-rich JPA implementations and offers object caching, statement caching, extra association mappings, and other features that help to improve performance and usefulness. Plus, Hibernate is open-source (with support offered by JBoss).
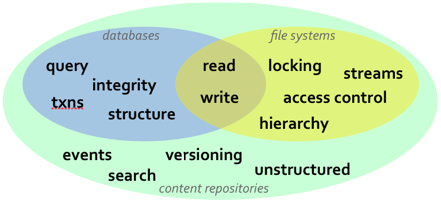
While relational databases and JPA are solutions that work well for many applications, they are more limited in cases when the information structure is highly flexible, the structure is not known a priori, or that structure is subject to frequent change and customization. In these situations, content repositories may offer a better choice for persistence. Content repositories are almost a hybrid with the storage capabilities of relational databases and the flexibility offered by other systems, such as using files. Content repositories also typically provide other capabilities as well, including versioning, indexing, search, access control, transactions, and observation. Because of this, content repositories are used by content management systems (CMS), document management systems (DMS), and other applications that manage electronic files (e.g., documents, images, multi-media, web content, etc.) and metadata associated with them (e.g., author, date, status, security information, etc.). The Content Repository for Java technology API provides a standard Java API for working with content repositories. Abbreviated "JCR", this API was developed as part of the Java Community Process under JSR-170 and is being revised under JSR-283.
The JCR API provides a number of information services that are needed by many applications, including: read and write access to information; the ability to structure information in a hierarchical and flexible manner that can adapt and evolve over time; ability to work with unstructured content; ability to (transparently) handle large strings; notifications of changes in the information; search and query; versioning of information; access control; integrity constraints; participation within distributed transactions; explicit locking of content; and of course persistence.
What makes JCR interesting, however, is that a JCR implementation provides all these features and capabilities regardless of where or how that information is persisted or stored. This is in fact the main purpose of JBoss DNA: provide a JCR implementation that provides access to content stored in many different kinds of systems. A JBoss DNA repository isn't yet another silo of information, but rather it's a JCR view of the information you already have in your environment: files systems, databases, other repositories, services, applications, etc. JBoss DNA can help you understand the systems and information you already have.
Of course when you start providing a unified view of all this information, you start recognizing the need to store more information, including metadata about and relationships between the existing content. JBoss DNA lets you do this, too. And JBoss DNA even tries to help you discover more about the information you already have, especially the information wrapped up in the kinds of files often found in enterprise systems: service definitions, policy files, images, media, documents, presentations, application components, reusable libraries, configuration files, application installations, databases schemas, management scripts, and so on. As files are loaded into the repository, JBoss DNA can sequence these files to extract from their content meaningful information that can be stored in the repository, where it can then be searched, accessed, and analyzed using the JCR API.
As we'll see in the next chapter, the ability of JBoss DNA to
federate, integrate, and sequence information make JBoss DNA a powerful asset and tool.
Then Chapter 3 will show that once a JBoss DNA repository is set up, application see
JBoss DNA just as another JCR javax.jcr.Repository instance and uses the standard JCR API to obtain a javax.jcr.Session
and work with the content.
Chapter 4 walks you through downloading and building the JBoss DNA examples, while Chapter 5 and Chapter 6 will run these very simple examples and walking through code. Chapter 7 wraps things up with a discussion about the future of JBoss DNA and what you can do next to start using JBoss DNA in your own applications.
There are lots of ways to use JBoss DNA in your own applications, but this chapter attempts to show some representative scenarios that take advantage of JBoss DNA's support for the JCR API as well as the federation, integration, and sequencing capabilities.
In a SOA environment, one important component is a service registry that provides versioned storage of all the artifacts that describe the services, their capabilities/restrictions, and the policies that surround them. Service repositories contain information that define the services and their message models, ownership, availability, security requirements/abilities, auditing, funding, monitoring, provisioning, provenance, usage, discovery mechanism, configuration, documentation, relationships to other services, classification taxonomies, ontologies, and many other important aspects.
A JCR repository provides an excellent starting point for a service repository. The ability to store a wide range of content, ranging from structured information to documents, means that a JCR repository can offer the flexibility to manage and organize the information while maintaining the ability to adapt the structure and schema as needs evolve over time.
A service repository will contain lots of information represented in different forms, and it's important that the repository make it easy for users to quickly find what they need. Organization of the information (probably in multiple hierarchies and with tags) is important, but more important is the ability for users to use simple searching (or more advanced queries) to return ranked results that match the criteria. For search to be effective, it is important that the repository understand the different kinds of artifacts that are uploaded and the information they contain.
JCR repositories are naturally searchable and queryable, but also can be used to integrate a taxonomy (or folksonomic tags) with the content, allowing the same content to be presented in different hierarchical classifications. But JBoss DNA capabilities also offer a great advantage, since any file that is uploaded can be automatically sequenced and processed to extract information that's meaningful and useful but often locked up within the file. For example, when a WSDL file is uploaded, the appropriate sequencer(s) process the file and extract and stores in the repository the structured information describing the types, message structures, operations, port types, bindings, and services found within the WSDL file. When an XML Schema Document is uploaded, JBoss DNA can do the same for the schema's complex and simple types, element and attribute declarations, model groups, namespaces, imports, includes, annotations, etc. And JBoss DNA can do the same for the various policy files, resource declarations, documentation, presentations, ontologies, etc.
Integration with a management system can be done in a similar manner. A JBoss DNA connector could access the management system to discover the servers and enable auto-discovery of the services, and "tag" the services deployments with the lifecycle phase (dev, test, production, etc.).
By using JBoss DNA, a service repository could manage the wide range of artifacts required in a SOA or web-oriented architecture, yet be able to present a unified view of all service information.
Many enterprise environments include numerous databases and data services, yet there is often no single place where all these different assets are described or related. A data source/service repository could provide information about the many databases running within the enterprise as well as their documentation, schema history, availability, usage policies, current users of the data (including applications, ETL processes, reporting), geographic deployments and synchronization, and the provenance of the data.
Some of this information may actually be defined or controlled within the data sources themselves or within other systems. For example, the DDL scripts used to migrate the database schemas are (hopefully) stored in a version control system, and the databases themselves have the ability to describe their current schemas. Using JBoss DNA, the repository could use a connector to the version control system to expose the scripts, as well as connectors to the databases to expose (and cache) the current schema of the databases. The data repository would then be able to allow users to search over this information without even touching the underlying systems.
However, the power of a data repository is really the ability to capture the relationships that otherwise were only captured in people's heads or trapped in documents spread throughout the network. A data repository can capture the policies that dictate how each data source should be used (which are for development purposes, or QA/testing purposes, or which are production, and how are they all related), and it can integrate with management systems to provide information about availability and deployment. As web services are created to provide service-based access to the data in databases, the repository can be used to maintain the relationships between these data services and the underlying sources. Similarly, the repository can track how the databases are used by applications, ETL processes, and reports.
Many applications and libraries have configuration files that allow the users (or developers) to dictate the setup and behavior. Often this involves multiple files in a specific structure on the file system. But invalid or inopportune changes to these files sometimes corrupt the environment, but creating a more robust configuration management system is often way beyond the desired effort.
An embedded JBoss DNA repository can provide a more formal and flexible configuration system with little effort. JCR's event system allows the system to be notified when the configuration changes, and versioning can help guarantee the ability to revert back to a previous (valid) configuration. JBoss DNA connectors can be used to integrate the files on the file system into the configuration system, keeping it natural for those wanting to view and change the configuration via the files. JBoss DNA sequencers can even process the configuration files to extract a more structured view of the system. And because JBoss DNA can used with a minimal footprint, it provides the ability to manage and version the configuration with little overhead.
JBoss DNA can even be used to centralize the configuration definition for a clustered or distributed system. In this mode, the configuration is managed in a central repository that is remotely accessible by the application. When a process is started, it examines the repository and reads the content containing its configuration. The application can monitor the configuration for changes so that it can modify itself and its components. For larger deployments, a central "enterprise configuration" repository can house the configuration of different kinds of systems, and can even be managed and manipulated through JCR.
As we'll see in the next chapter, this is actually the way in which JBoss DNA manages its own configuration. In the embedded case, the configuration repository is simply a local (in-memory) repository that is populated by the configuration file (or programmatic API). In a clustered mode, the repository is centralized. But either way, to JBoss DNA the configuration is always defined in a repository.
The scenarios described in this chapter are representative of some of the ways in which JBoss DNA can be used, and hopefully give you ideas about how you can leverage JBoss DNA in your application or library.
In the next chapter, we'll show how you can set up JBoss DNA and use it via the standard JCR API.
Using JBoss DNA within your application is actually quite straightforward. As you'll see in this chapter,
the first step is setting up JBoss DNA and starting the JcrEngine. After that, you obtain the
javax.jcr.Repository instance for a named repository and just use the standard JCR API throughout your
application.
JBoss DNA encapsulates everything necessary to run one or more JCR repositories into a single JcrEngine instance.
This includes all underlying repository sources, the pools of connections to the sources, the sequencers,
the MIME type detector(s), and the Repository implementations.
Obtaining a JcrEngine instance is very easy - assuming that you have a valid JcrConfiguration instance. We'll see
how to get one of those in a little bit, but if you have one then all you have to do is build and start the engine:
JcrConfiguration config = ...
JcrEngine engine = config.build();
engine.start();
Obtaining a JCR Repository instance is a matter of simply asking the engine for it by the name defined in the configuration:
javax.jcr.Repository repository = engine.getRepository("Name of repository");
At this point, your application can proceed by working with the JCR API.
And, once you're finished with the JcrEngine, you should shut it down:
engine.shutdown();
engine.awaitTermination(3,TimeUnit.SECONDS); // optional
When the shutdown() method is called, the Repository instances managed by the engine are marked as being shut down,
and they will not be able to create new Sessions. However, any existing Sessions or ongoing operations (e.g., event notifications)
present at the time of the shutdown() call will be allowed to finish.
In essence, shutdown() is a graceful request, and since it may take some time to complete,
you can wait until the shutdown has completed by simply calling awaitTermination(...) as shown above.
This method will block until the engine has indeed shutdown or until the supplied time duration has passed (whichever comes first).
And, yes, you can call the awaitTermination(...) method repeatedly if needed.
The previous section assumed the existence of a JcrConfiguration. It's not really that creating an instance is all that difficult.
In fact, there's only one no-argument constructor, so actually creating the instance is a piece of cake. What can be a little more challenging,
though, is setting up the JcrConfiguration instance, which must define the following components:
Repository sourcesare the POJO objects that each describe a particular location where content is stored. Each repository source object is an instance of a JBoss DNA connector, and is configured with the properties that particular source. JBoss DNA's RepositorySource classes are analogous to JDBC'sDataSourceclasses - they are implemented by specific connectors (aka, "drivers") for specific kinds of repository sources (aka, "databases"). Similarly, a RepositorySource instance is analogous to aDataSourceinstance, with bean properties for each configurable parameter. Therefore, each repository source definition must supply the name of the RepositorySource class, any bean properties, and, optionally, the classpath that should be used to load the class.Repositoriesdefine the JCR repositories that are available. Each repository has a unique name that is used to obtain the Repository instance from theJcrEngine'sgetRepository(String)method, but each repository definition also can include the predefined namespaces (other than those automatically defined by JBoss DNA), various options, and the node types that are to be available in the repository without explicit registration through the JCR API.Sequencersdefine the particular sequencers that are available for use. Each sequencer definition provides the path expressions governing which nodes in the repository should be sequenced when those nodes change, and where the resulting output generated by the sequencer should be placed. The definition also must state the name of the sequencer class, any bean properties and, optionally, the classpath that should be used to load the class.MIME type detectorsdefine the particular MIME type detector(s) that should be made available. A MIME type detector does exactly what the name implies: it attempts to determine the MIME type given a "filename" and contents. JBoss DNA automatically uses a detector that uses the file extension to identify the MIME type, but also provides an implementation that uses an external library to identify the MIME type based upon the contents. The definition must state the name of the detector class, any bean properties and, optionally, the classpath that should be used to load the class.
There really are three options:
Load from a fileis conceptually the easiest and requires the least amount of Java code, but it now requires a configuration file.Load from a configuration repositoryis not much more complicated than loading from a file, but it does allow multipleJcrEngineinstances (usually in different processes perhaps on different machines) to easily access their (shared) configuration. And technically, loading the configuration from a file really just creates anInMemoryRepositorySource, imports the configuration file into that source, and then proceeds with this approach.Programmatic configurationis always possible, even if the configuration is loaded from a file or repository. Using theJcrConfiguration's API, you can define (or update or remove) all of the definitions that make up a configuration.
Each of these approaches has their obvious advantages, so the choice of which one to use is entirely up to you.
Loading the JBoss DNA configuration from a file is actually very simple:
JcrConfiguration config = new JcrConfiguration();
configuration.loadFrom(file);
where the file parameter can actually be a File instance, a URL to the file, an InputStream
containing the contents of the file, or even a String containing the contents of the file.
Note
The loadFrom(...) method can be called any number of times, but each time it is called it completely wipes
out any current notion of the configuration and replaces it with the configuration found in the file.
There is an optional second parameter that defines the Path within the configuration file identifying the parent node of the various configuration nodes. If not specified, it assumes "/". This makes it possible for the configuration content to be located at a different location in the hierarchical structure. (This is not often required, but when it is required this second parameter is very useful.)
Here is the configuration file that is used in the repository example, though it has been simplified a bit and most comments have been removed for clarity):
<?xml version="1.0" encoding="UTF-8"?>
<configuration xmlns="http://www.jboss.org/dna/1.0" xmlns:jcr="http://www.jcp.org/jcr/1.0">
<!--
Define the JCR repositories
-->
<dna:repositories>
<!--
Define a JCR repository that accesses the 'Cars' source directly.
This of course is optional, since we could access the same content through 'vehicles'.
-->
<dna:repository jcr:name="car repository" dna:source="Cars">
<dna:options jcr:primaryType="dna:options">
<jaasLoginConfigName jcr:primaryType="dna:option" dna:value="dna-jcr"/>
</dna:options>
</dna:repository>
</dna:repositories>
<!--
Define the sources for the content. These sources are directly accessible using the DNA-specific Graph API.
-->
<dna:sources jcr:primaryType="nt:unstructured">
<dna:source jcr:name="Cars" dna:classname="org.jboss.dna.graph.connector.inmemory.InMemoryRepositorySource" dna:retryLimit="3" dna:defaultWorkspaceName="workspace1"/>
<dna:source jcr:name="Aircraft" dna:classname="org.jboss.dna.graph.connector.inmemory.InMemoryRepositorySource">
<!-- Define the name of the workspace used by default. Optional, but convenient. -->
<defaultWorkspaceName>workspace2</defaultWorkspaceName>
</dna:source>
</dna:sources>
<!--
Define the sequencers. This is an optional section. For this example, we're not using any sequencers.
-->
<dna:sequencers>
<!--dna:sequencer jcr:name="Image Sequencer" dna:classname="org.jboss.dna.sequencer.image.ImageMetadataSequencer">
<dna:description>Image metadata sequencer</dna:description>
<dna:pathExpression>/foo/source => /foo/target</dna:pathExpression>
<dna:pathExpression>/bar/source => /bar/target</dna:pathExpression>
</dna:sequencer-->
</dna:sequencers>
<dna:mimeTypeDetectors>
<dna:mimeTypeDetector jcr:name="Detector" dna:description="Standard extension-based MIME type detector"/>
</dna:mimeTypeDetectors>
</configuration>
Loading the JBoss DNA configuration from an existing repository is also pretty straightforward. Simply create and configure the
RepositorySource instance to point to the desired repository, and then call the loadFrom(RepositorySource source)
method:
RepositorySource configSource = ...
JcrConfiguration config = new JcrConfiguration();
configuration.loadFrom(configSource);
This really is a more advanced way to define your configuration, so we won't go into how you configure a RepositorySource. For more information, consult the Reference Guide.
Note
The loadFrom(...) method can be called any number of times, but each time it is called it completely wipes
out any current notion of the configuration and replaces it with the configuration found in the file.
There is an optional second parameter that defines the name of the workspace in the supplied source where the configuration content can be found. It is not needed if the workspace is the source's default workspace. There is an optional third parameter that defines the Path within the configuration repository identifying the parent node of the various configuration nodes. If not specified, it assumes "/". This makes it possible for the configuration content to be located at a different location in the hierarchical structure. (This is not often required, but when it is required this second parameter is very useful.)
Defining the configuration programmatically is not terribly complicated, and it for obvious reasons results in more verbose Java code. But this approach is very useful and often the easiest approach when the configuration must change or is a reflection of other dynamic information.
The JcrConfiguration class was designed to have an easy-to-use API that makes it easy to configure each of the different kinds of
components, especially when using an IDE with code completion. Here are several examples:
Each repository source definition must include the name of the RepositorySource class as well as each bean property that should be set on the object:
JcrConfiguration config = ...
config.repositorySource("source A")
.usingClass(InMemoryRepositorySource.class)
.setDescription("The repository for our content")
.setProperty("defaultWorkspaceName", workspaceName);
This example defines an in-memory source with the name "source A", a description, and a single "defaultWorkspaceName" bean property. Different RepositorySource implementations will the bean properties that are required and optional. Of course, the class can be specified as Class reference or a string (followed by whether the class should be loaded from the classpath or from a specific classpath).
Note
Each time repositorySource(String) is called, it will either load the existing definition with the supplied
name or will create a new definition if one does not already exist. To remove a definition, simply call remove()
on the result of repositorySource(String).
The set of existing definitions can be accessed with the repositorySources() method.
Each repository must be defined to use a named repository source, but all other aspects (e.g., namespaces, node types, options) are optional.
JcrConfiguration config = ...
config.repository("repository A")
.addNodeTypes("myCustomNodeTypes.cnd")
.setSource("source 1")
.registerNamespace("acme","http://www.example.com/acme")
.setOption(JcrRepository.Option.JAAS_LOGIN_CONFIG_NAME, "dna-jcr");
This example defines a repository that uses the "source 1" repository source (which could be a federated source, an in-memory source, a database store, or any other source). Additionally, this example adds the node types in the "myCustomNodeTypes.cnd" file as those that will be made available when the repository is accessed. It also defines the "http://www.example.com/acme" namespace, and finally sets the "JAAS_LOGIN_CONFIG_NAME" option to define the name of the JAAS login configuration that should be used by the JBoss DNA repository.
Note
Each time repository(String) is called, it will either load the existing definition with the supplied
name or will create a new definition if one does not already exist. To remove a definition, simply call remove()
on the result of repository(String).
The set of existing definitions can be accessed with the repositories() method.
Each defined sequencer must specify the name of the StreamSequencer implementation class as well as the path expressions defining which nodes should be sequenced and the output paths defining where the sequencer output should be placed (often as a function of the input path expression).
JcrConfiguration config = ...
config.sequencer("Image Sequencer")
.usingClass("org.jboss.dna.sequencer.image.ImageMetadataSequencer")
.loadedFromClasspath()
.setDescription("Sequences image files to extract the characteristics of the image")
.sequencingFrom("//(*.(jpg|jpeg|gif|bmp|pcx|png|iff|ras|pbm|pgm|ppm|psd)[*])/jcr:content[@jcr:data]")
.andOutputtingTo("/images/$1");
This shows an example of a sequencer definition named "Image Sequencer" that uses the ImageMetadataSequencer class
(loaded from the classpath), that is to sequence the "jcr:data" property on any new or changed nodes that are named
"jcr:content" below a parent node with a name ending in ".jpg", ".jpeg", ".gif", ".bmp", ".pcx", ".iff", ".ras",
".pbm", ".pgm", ".ppm" or ".psd". The output of the sequencing operation should be placed at the "/images/$1" node,
where the "$1" value is captured as the name of the parent node. (The capture groups work the same was as regular expressions;
see the Reference Guide for more details.)
Of course, the class can be specified as Class reference or a string (followed by whether the class should be loaded from
the classpath or from a specific classpath).
Note
Each time sequencer(String) is called, it will either load the existing definition with the supplied
name or will create a new definition if one does not already exist. To remove a definition, simply call remove()
on the result of sequencer(String).
The set of existing definitions can be accessed with the sequencers() method.
Each defined MIME type detector must specify the name of the MimeTypeDetector implementation class as well as any other bean properties required by the implementation.
JcrConfiguration config = ...
config.mimeTypeDetector("Extension Detector")
.usingClass(org.jboss.dna.graph.mimetype.ExtensionBasedMimeTypeDetector.class);
Of course, the class can be specified as Class reference or a string (followed by whether the class should be loaded from the classpath or from a specific classpath).
Note
Each time mimeTypeDetector(String) is called, it will either load the existing definition with the supplied
name or will create a new definition if one does not already exist. To remove a definition, simply call remove()
on the result of mimeTypeDetector(String).
The set of existing definitions can be accessed with the mimeTypeDetectors() method.
This chapter outline how you configure JBoss DNA, how you then access a javax.jcr.Repository instance,
and use the standard JCR API to interact with the repository. The
next chapter walks you through downloading
and running the JBoss DNA examples.
This chapter provides instructions for downloading and running a sample application that demonstrates how JBoss DNA works with a JCR repository to automatically sequence changing content to extract useful information. So read on to get the simple application running.
JBoss DNA uses Maven 2 for its build system, as is this example. Using Maven 2 has several advantages, including the ability to manage dependencies. If a library is needed, Maven automatically finds and downloads that library, plus everything that library needs. This means that it's very easy to build the examples - or even create a maven project that depends on the JBoss DNA JARs.
Note
To use Maven with JBoss DNA, you'll need to have JDK 5 or 6 and Maven 2.0.9 (or higher).
Maven can be downloaded from http://maven.apache.org/, and is installed by unzipping the
maven-2.0.9-bin.zip file to a convenient location on your local disk. Simply add $MAVEN_HOME/bin
to your path and add the following profile to your ~/.m2/settings.xml file:
<settings>
<profiles>
<profile>
<id>jboss.repository</id>
<activation>
<property>
<name>!jboss.repository.off</name>
</property>
</activation>
<repositories>
<repository>
<id>snapshots.jboss.org</id>
<url>http://snapshots.jboss.org/maven2</url>
<snapshots>
<enabled>true</enabled>
</snapshots>
</repository>
<repository>
<id>repository.jboss.org</id>
<url>http://repository.jboss.org/maven2</url>
<snapshots>
<enabled>false</enabled>
</snapshots>
</repository>
</repositories>
<pluginRepositories>
<pluginRepository>
<id>repository.jboss.org</id>
<url>http://repository.jboss.org/maven2</url>
<snapshots>
<enabled>false</enabled>
</snapshots>
</pluginRepository>
<pluginRepository>
<id>snapshots.jboss.org</id>
<url>http://snapshots.jboss.org/maven2</url>
<snapshots>
<enabled>true</enabled>
</snapshots>
</pluginRepository>
</pluginRepositories>
</profile>
</profiles>
</settings>
This profile informs Maven of the two JBoss repositories (snapshots and releases) that contain all of the JARs for JBoss DNA and all dependent libraries.
The next step is to download the example for this Getting Started guide, and extract the contents to a convenient location on your local disk. You'll find the example contains the following files, which are organized according to the standard Maven directory structure:
examples/pom.xml
sequencers/pom.xml
/src/main/assembly
/config
/java
/resources
/test/java
/resources
repository/pom.xml
/src/main/assembly
/config
/java
/resources
/test/java
/resources
There are essentially three Maven projects: a sequencers project, a repository project,
and a parent project. All of the source for the sequencing example is located in the sequencers subdirectory,
while all of the source for the repository example is located in the repository subdirectory.
And you may have noticed that none of the JBoss DNA libraries are there. This is where Maven comes in.
The two pom.xml files tell Maven everything it needs to know about what libraries are required and
how to build the example.
In a terminal, go to the examples directory and run:
$ mvn install
This command downloads all of the JARs necessary to compile and build the example, including the JBoss DNA libraries, the libraries they depend on, and any missing Maven components. (These are downloaded from the JBoss repositories only once and saved on your machine. This means that the next time you run Maven, all the libraries will already be available locally, and the build will run much faster.) The command then continues by compiling the example's source code (and unit tests) and running the unit tests. The build is successful if you see the following:
$ mvn install ... [INFO] ------------------------------------------------------------------------ [INFO] Reactor Summary: [INFO] ------------------------------------------------------------------------ [INFO] Getting Started examples .............................. SUCCESS [2.106s] [INFO] Sequencer Examples .................................... SUCCESS [9.768s] [INFO] ------------------------------------------------------------------------ [INFO] ------------------------------------------------------------------------ [INFO] BUILD SUCCESSFUL [INFO] ------------------------------------------------------------------------ [INFO] Total time: 12 seconds [INFO] Finished at: Wed May 07 12:00:06 CDT 2008 [INFO] Final Memory: 14M/28M [INFO] ------------------------------------------------------------------------ $
If there are errors, check whether you have the correct version of Maven installed and that you've correctly updated your Maven settings as described above.
If you've successfully built the examples, there will be a new examples/sequencers/target/ directory that contains
all of the generated output for the sequencers example, including a dna-example-sequencers-basic.dir/ subdirectory
that contains the following:
run.shis the *nix shell script that will run the sequencer example application.log4j.propertiesis the Log4J configuration file.sample1.mp3is a sample MP3 audio file you'll use later to upload into the repository.caution.gif,caution.png,caution.jpg, andcaution.pictare images that you'll use later and upload into the repository.sequencing.cndis a Compact Node Definition (CND) file that defines the node types used in the output from the sequencers.securitysubdirectory containing several files related to the JAAS implementation used for authentication.project1subdirectory contains some Java source that can be loaded into the repository.libsubdirectory contains the JARs for all of the JBoss DNA artifacts as well as those for other libraries required by JBoss DNA and the sequencer example.
Similarly, the examples/repository/target/ directory contains all of the generated output for the repository example, including
a dna-example-repository-basic.dir/ subdirectory that contains the following:
run.shis the *nix shell script that will run the repository example application.log4j.propertiesis the Log4J configuration file.aircraft.xmlis an XML file containing the information that the example application imports into its "Aircraft" repository.cars.xmlis an XML file containing the information that the example application imports into its "Cars" repository.aircraft.cnd,cars.cnd, andvehicles.cndare the CND files used for the three different JCR Repositories set up in the example. Thevehicles.cndis just a combination of the other two (with duplicates removed).configRepository.xmlis an XML file containing the information that the example application loads as its configuration and which defines the sources, repositories, sequencers (if used), and other components that make up the DNA JCR engine.securitysubdirectory containing several files related to the JAAS implementation used for authentication.libsubdirectory contains the JARs for all of the JBoss DNA artifacts as well as those for other libraries required by JBoss DNA and the repository example.
The previous chapter walked through the process of downloading and building the examples. This chapter will focus on the sequencer example, showing how to run the example and then walking through the code to describe what it's doing.
The sequencing example consists of a client application that sets up an in-memory JCR repository and that allows a user to upload files into that repository. The client also sets up the DNA services with two sequencers so that if any of the uploaded files are PNG, JPEG, GIF, BMP or other images, DNA will automatically extract the image's metadata (e.g., image format, physical size, pixel density, etc.) and store that in the repository. Alternatively, if the uploaded file is an MP3 audio file, DNA will extract some of the ID3 metadata (e.g., the author, title, album, year and comment) and store that in the repository.
To run the client application, go to the examples/sequencers/target/dna-example-sequencers-basic.dir/
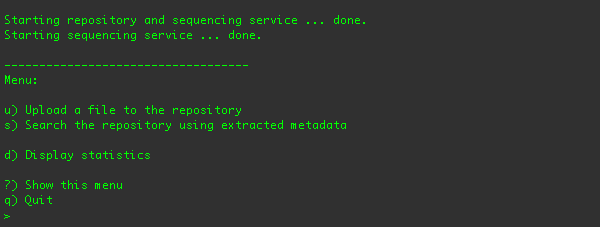
directory and type ./run.sh. You should see the command-line client and its menus in your terminal:
From this menu, you can upload a file into the repository, search for media in the repository, print sequencing statistics,
or quit the application.
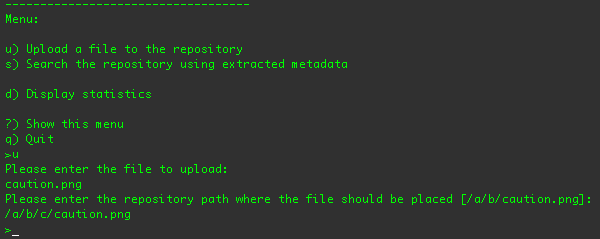
The first step is to upload one of the example images. If you type 'u' and press return, you'll be prompted to supply the
path to the file you want to upload. Since the application is running from within the
examples/sequencers/target/dna-example-sequencers-basic.dir/ directory, you can specify any of the files
in that directory without specifying the path:
You can specify any fully-qualified or relative path. The application will notify you if it cannot find the file you
specified. The example client configures JBoss DNA to sequence MP3 audio files, Java source files, or image files with one of
the following extensions (technically, nodes that have names ending in the following):
jpg, jpeg, gif, bmp, pcx, png,
iff, ras, pbm, pgm, ppm, and psd.
Files with other extensions in the repository path will be ignored. For your convenience, the example provides several
files that will be sequenced (caution.png, caution.jpg, caution.gif, and
sample1.mp3) and one image that will not be sequenced (caution.pict). Feel free to try other files.
After you have specified the file you want to upload, the example application asks you where in the repository you'd like to
place the file. (If you want to use the suggested location, just press return.) The client application
uses the JCR API to upload the file to that location in the repository, creating any nodes (of type nt:folder)
for any directories that don't exist, and creating a node (of type nt:file) for the file. And, per the JCR specification,
the application creates a jcr:content node (of type nt:resource) under the file node.
The file contents are placed on this jcr:content node in the jcr:data property. For example, if you specify
/a/b/caution.png, the following structure will be created in the repository:
/a (nt:folder)
/b (nt:folder)
/caution.png (nt:file)
/jcr:content (nt:resource)
@jcr:data = {contents of the file}
@jcr:mimeType = {mime type of the file}
@jcr:lastModified = {now}
Other kinds of files are treated in a similar way.
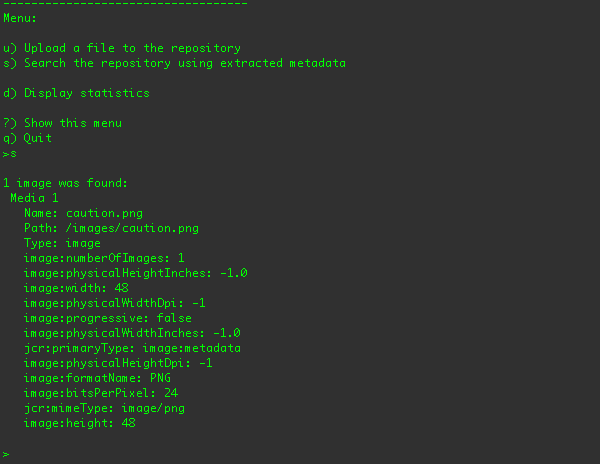
When the client uploads the file using the JCR API, DNA gets notified of the changes, consults the sequencers to see whether
any of them are interested in the new or updated content, and if so runs those sequencers. The image sequencer processes image
files for metadata, and any metadata found is stored under the /images branch of the repository. The MP3 sequencer
processes MP3 audio files for metadata, and any metadata found is stored under the /mp3s
branch of the repository. And metadata about Java classes are stored under the /java area of the repository.
All of this happens asynchronously, so any DNA activity doesn't impede or slow down the client activities.
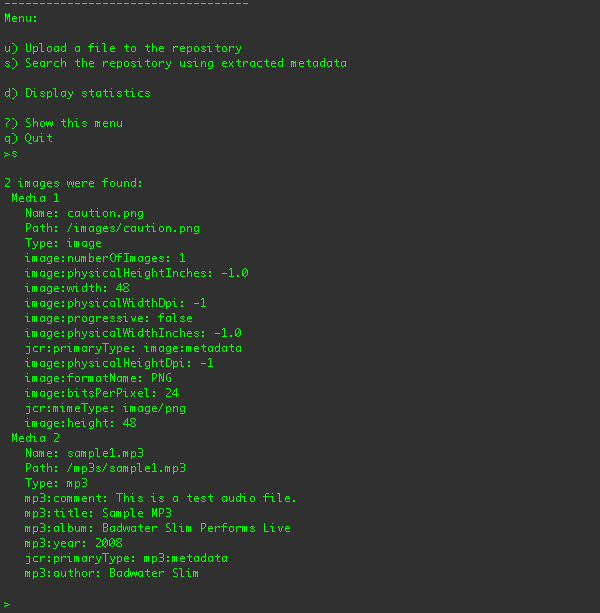
So, after the file is uploaded, you can search the repository for the image metadata using the "s" menu option:
Here are the search results after the sample1.mp3 audio file has been uploaded (to the /a/b/sample1.mp3 location):
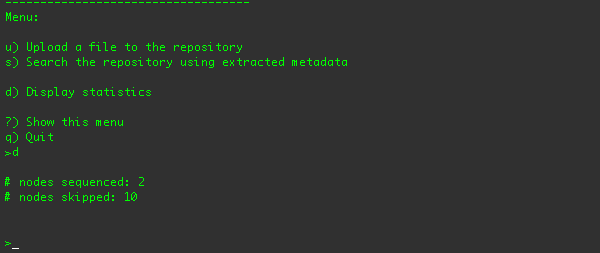
You can also display the sequencing statistics using the "d" menu option:
These stats show how many nodes were sequenced, and how many nodes were skipped because they didn't apply to the sequencer's
criteria.
Note
There will probably be more nodes skipped than sequenced, since there are more nt:folder and nt:resource
nodes than there are nt:file nodes with acceptable names.
You can repeat this process with other files. Any file that isn't an image or MP3 files (as recognized by the sequencing configurations that we'll describe later) will not be sequenced.
Recall that the example application consists of a client application that sets up an in-memory JCR repository and that allows a user to upload files into that repository. The client also sets up the DNA services with an image sequencer so that if any of the uploaded files are PNG, JPEG, GIF, BMP or other images, DNA will automatically extract the image's metadata (e.g., image format, physical size, pixel density, etc.) and store that in the repository. Or, if the client uploads MP3 audio files, the title, author, album, year, and comment are extracted from the audio file and stored in the repository.
The example is comprised of 5 classes and 1 interface, located in the src/main/java directory:
org/jboss/example/dna/sequencers/ConsoleInput.java
/ContentInfo.java
/JavaInfo.java
/MediaInfo.java
/SequencingClient.java
/UserInterface.java
SequencingClient is the class that contains the main application. ContentInfo is a simple class
that encapsulate metadata generated by the sequencers and accessed by this example application, and there are two subclasses:
MediaInfo encapsulates metadata about media (image and MP3) files, while JavaInfo is a subclass
encapsulating information about a Java class. The client accesses the content from the repository and represent the
information using instances of ContentInfo (and its subclasses) and then passing them to the UserInterface.
UserInterface is an interface with methods that will be called at runtime to
request data from the user. ConsoleInput is an implementation of this that creates a text user interface,
allowing the user to operate the client from the command-line. We can easily create a graphical implementation of
UserInterface at a later date. We can also create a mock implementation for testing purposes that simulates
a user entering data. This allows us to check the behavior of the client automatically using conventional JUnit test cases,
as demonstrated by the code in the src/test/java directory:
org/jboss/example/dna/sequencers/SequencingClientTest.java
/MockUserInterface.java
If we look at the SequencingClient code, there are a handful of methods that encapsulate the various activities.
Note
Some of the code samples included in this book have had some of the error handling and comments removed so that the code is more readable and concise.
The main(String[] argv) method is of course the method that is executed when the application is run. This code
creates the JBoss DNA configuration using the programmatic style.
// Create the configuration.
String repositoryId = "content";
String workspaceName = "default";
JcrConfiguration config = new JcrConfiguration();
// Set up the in-memory source where we'll upload the content and where the sequenced output will be stored ...
config.repositorySource("store")
.usingClass(InMemoryRepositorySource.class)
.setDescription("The repository for our content")
.setProperty("defaultWorkspaceName", workspaceName);
// Set up the JCR repository to use the source ...
config.repository(repositoryId)
.addNodeTypes("sequencing.cnd")
.setSource("store");
// Set up the image sequencer ...
config.sequencer("Image Sequencer")
.usingClass("org.jboss.dna.sequencer.image.ImageMetadataSequencer")
.loadedFromClasspath()
.setDescription("Sequences image files to extract the characteristics of the image")
.sequencingFrom("//(*.(jpg|jpeg|gif|bmp|pcx|png|iff|ras|pbm|pgm|ppm|psd)[*])/jcr:content[@jcr:data]")
.andOutputtingTo("/images/$1");
// Set up the MP3 sequencer ...
config.sequencer("MP3 Sequencer")
.usingClass("org.jboss.dna.sequencer.mp3.Mp3MetadataSequencer")
.loadedFromClasspath()
.setDescription("Sequences mp3 files to extract the id3 tags of the audio file")
.sequencingFrom("//(*.mp3[*])/jcr:content[@jcr:data]")
.andOutputtingTo("/mp3s/$1");
// Set up the Java source file sequencer ...
config.sequencer("Java Sequencer")
.usingClass("org.jboss.dna.sequencer.java.JavaMetadataSequencer")
.loadedFromClasspath()
.setDescription("Sequences Java files to extract the AST structure of the Java source code")
.sequencingFrom("//(*.java[*])/jcr:content[@jcr:data]")
.andOutputtingTo("/java/$1");
// Now start the client and tell it which repository and workspace to use ...
SequencingClient client = new SequencingClient(config, repositoryId, workspaceName);
client.setUserInterface(new ConsoleInput(client));
The first block of code configures the JcrConfiguration and sets up the "store" source, the "content" repository,
and three sequencers. Again, this is done via the programmatic style. An alternative would be to load the entire configuration
from a configuration file or from an existing configuration repository. (The repository example shown in the
next chapter shows how to load the configuration from a file.)
The second block simply instantiates the SequencingClient class, passing the configuration and the name of the repository
and workspace, and finally sets the user interface (which then executes its behavior, which we'll see below).
The startRepository() method builds the JcrEngine component from the configuration, starts the engine,
and obtains the JCR javax.jcr.Repository instance that the client will use. Note that the client has not yet
obtained a javax.jcr.Session instance, since this will be done each time the client needs to access content from
the repository. (This is actually a common practice according to the JCR specification, since Sessions are intended to be
very lightweight.)
public void startRepository() throws Exception {
if (this.repository == null) {
try {
// Start the DNA engine ...
this.engine = this.configuration.build();
this.engine.start();
// Now get the JCR repository instance ...
this.repository = this.engine.getRepository(repositoryName);
} catch (Exception e) {
this.repository = null;
throw e;
}
}
}
The shutdownRepository() method requests the JcrEngine instance shuts down and, since that may take
a few moments (if there are any ongoing operations or enqueued activities) awaits for it to complete the shutdown.
public void shutdownRepository() throws Exception {
if (this.repository != null) {
try {
this.engine.shutdown();
this.engine.awaitTermination(4, TimeUnit.SECONDS);
} finally {
this.repository = null;
}
}
}
None of the other methods really do anything with JBoss DNA per se. Instead, they merely work with the repository using the JCR API.
If we look at the ConsoleInput constructor, it starts the repository and a thread
for the user interface. At this point, the constructor returns, but the main application continues under the user interface thread.
When the user requests to quit, the user interface thread also shuts down the JCR repository.
public ConsoleInput( SequencerClient client ) {
try {
client.startRepository();
System.out.println(getMenu());
Thread eventThread = new Thread(new Runnable() {
private boolean quit = false;
public void run() {
try {
while (!quit) {
// Display the prompt and process the requested operation ...
}
} finally {
try {
// Terminate ...
client.shutdownRepository();
} catch (Exception err) {
System.out.println("Error shutting down sequencing service and repository: "
+ err.getLocalizedMessage());
err.printStackTrace(System.err);
}
}
}
});
eventThread.start();
} catch (Exception err) {
System.out.println("Error: " + err.getLocalizedMessage());
err.printStackTrace(System.err);
}
}
There is one more aspect of this example that is worth discussing. While the repository example in the
next chapter does show how to use JAAS, this example intentionally shows how
you might integrate a different security system into JBoss DNA. In the createSession() method,
the RepositoryClient creates a SecurityContextCredentials wrapper around a custom
SecurityContext implementation, then passes that credentials into the login(Credentials,String)
method:
protected Session createSession() throws RepositoryException {
SecurityContext securityContext = new MyCustomSecurityContext();
SecurityContextCredentials credentials = new SecurityContextCredentials(securityContext);
return this.repository.login(credentials, workspaceName);
}
where the custom SecurityContext implementation is as follows:
protected class MyCustomSecurityContext implements SecurityContext {
/**
* @see org.jboss.dna.graph.SecurityContext#getUserName()
*/
public String getUserName() {
return "Fred";
}
/**
* @see org.jboss.dna.graph.SecurityContext#hasRole(java.lang.String)
*/
public boolean hasRole( String roleName ) {
return true;
}
/**
* @see org.jboss.dna.graph.SecurityContext#logout()
*/
public void logout() {
// do something
}
}
Obviously you would want to implement this correctly. If you're using JBoss DNA in a web application, your SecurityContext
implementation would likely delegate to the HttpServletRequest.
But if you're using JAAS, then you could just pass in a javax.jcr.SimpleCredentials with the username and password,
as long as your JcrConfiguration's repository definitions are set up to use the correct JAAS login context name
(see the repository example in the next chapter). Or, you could use the approach listed
above and supply an instance of the JaasSecurityContext to the SecurityContextCredentials.
At this point, we've reviewed all of the interesting code in the example application related to JBoss DNA. However, feel free to play with the application, trying different things.
This chapter walked through running the sequencer example and looked at the example code. With the sequencer client, you could upload files into a JCR repository, while JBoss DNA automatically sequenced the image, MP3, or Java source files you uploaded, extracted the metadata from the files, and stored that metadata inside the repository.
In the next chapter we'll do the same for the repository example.
Chapter 4 walked through the process of downloading and building the examples, while the previous chapter showed how to run the sequencer example and walked through the code. In this chapter, we'll run the repository example and walk through that example code to see what it's doing.
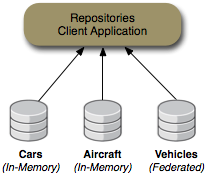
The repository example consists of a client application that sets up two DNA repositories (named "Cars" and "Airplanes") and a federated repository ("Vehicles") that dynamically federates the information from the other two repositories. The client application allows you to interactively navigate each of these repositories just as you would navigate the directory structure on a file system.
This collection of repositories is shown in the following figure:
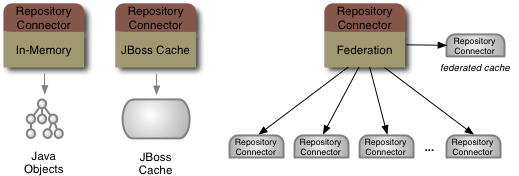
Most of the repositories are in-memory repositories (using the In-Memory repository connector), but the federated "Vehicles" repository
content is federated from the other repositories and cached into the "Cache" repository. This is shown in the following figure:
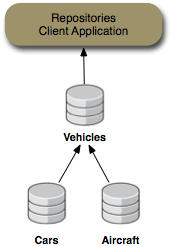
Figure 6.2. Vehicles repository content is federated from the Cars, Airplanes and Configuration repositories
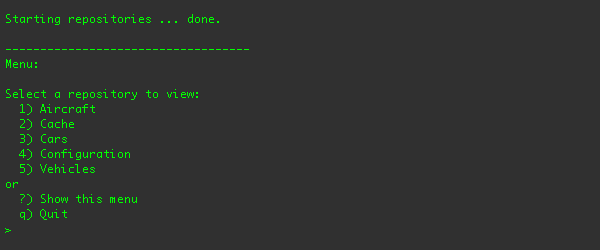
To run the client application, go to the examples/repository/target/dna-example-repositories-basic.dir/
directory and type ./run.sh. You should see the command-line client and its menus in your terminal:
From this menu, you can see the list of repositories, select one, and navigate through that repository in a manner similar
to a *nix command-line shell (although the client itself uses the JCR API to interact with the repositories).
Here are some of the commands you can use:
Table 6.1. Repository client commands to navigate a repository
| Command | Description |
|---|---|
| pwd | Print the path of the current node (e.g., the "working directory") |
| ls [path] | List the children and properties of the node at the supplied path, where "path" can be any relative path or absolute path. If "path" is not supplied, the current working node's path is used. |
| cd path | Change to the specified node, where "path"
can be any relative path or absolute path. For example, "cd alpha" changes the current node to be a child named
"alpha"; "cd .." changes the current node to the parent node; "cd /a/b" changes
the current node to be the "/a/b" node. |
| exit | Exit this repository and return the list of repositories. |
Note
The first time you access any repository, you will be prompted to log in. This is JAAS at work, and the example application should prompt you for a username and password. As configured in the "jaas.conf.xml" and "users.properties" files, enter "jsmith" for the username and "secret" for the password. And, yes, the password is shown in cleartext - this is a simple example, after all!
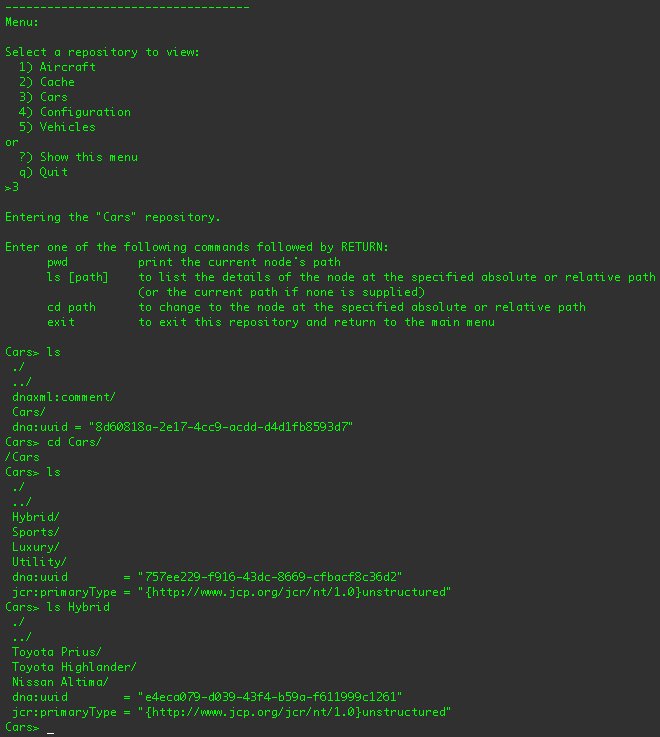
If you were to select the "Cars" repository and use some of the commands, you should see something similar to:
You can also choose to navigate the "Vehicles" repository, which projects the "Cars" repository content under the
/Vehicles/Cars node, the "Airplanes" content under the /Vehicles/Airplanes branch,
and the "Configuration" content under /dna:system.
Try using the client to walk the different repositories. And while this is a contrived application, it does demonstrate the use of JBoss DNA to federate repositories and provide access through JCR.
As mentioned in the Introduction, one of the capabilities of JBoss DNA is to provide access through JCR to different kinds of repositories and storage systems. Your applications work with the JCR API, but through JBoss DNA you're able to accesses the content from where the information exists - not just a single purpose-built repository. This is fundamentally what makes JBoss DNA different.
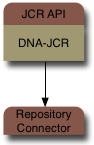
How does JBoss DNA do this? At the heart of JBoss DNA and it's JCR implementation is a simple connector system that is designed around creating and accessing graphs. The JBoss DNA JCR implementation actually just sits on top of a single repository source, which it uses to access of the repositories content.
That single repository source could be an in-memory repository, a JBoss Cache instance, or a federated repository.
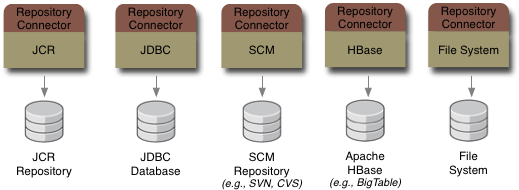
And the JBoss DNA project has plans to create other connectors, too. For instance, we're going to build a connector
to other JCR repositories. And another to a file system, so that the files and directories on an area of the file system
can be accessed through JCR. Of course, if we don't have a connector to suit your needs, you can write your own.
Note
You might be thinking that these connectors are interesting, but what do they really provide? Is it really useful to use JCR to access a relational database rather than JDBC? Or, why access the files on a file system when there are already mechanisms to do that?
Maybe putting JCR on top of a single system (like a JDBC database) isn't that interesting. What is interesting, though, is accessing the information in multiple systems as if all that information were in a single JCR repository. That's what the federated repository source is all about. The JBoss DNA connector system just makes it possible to interact with all these systems in the same way.
Think of it this way: with JBoss DNA, you can use JCR to get to the schemas of multiple relational databases and the schemas defined by DDL files in your SVN repository and the schemas defined by logical models stored on your file system.
Before we go further, let's define some terminology regarding connectors.
A connector is the runnable code packaged in one or more JAR files that contains implementations of several interfaces (described below). A Java developer writes a connector to a type of source, such as a particular database management system, LDAP directory, source code management system, etc. It is then packaged into one or more JAR files (including dependent JARs) and deployed for use in applications that use JBoss DNA repositories.
The description of a particular source system (e.g., the "Customer" database, or the company LDAP system) is called a repository source. JBoss DNA defines a RepositorySource interface that defines methods describing the behavior and supported features and a method for establishing connections. A connector will have a class that implements this interface and that has JavaBean properties for all of the connector-specific properties required to fully describe an instance of the system. Use of JavaBean properties is not required, but it is highly recommended, as it enables reflective configuration and administration. Applications that use JBoss DNA create an instance of the connector's RepositorySource implementation and set the properties for the external source that the application wants to access with that connector.
A repository source instance is then used to establish connections to that source. A connector provides an implementation of the RepositoryConnection interface, which defines methods for interacting with the external system. In particular, the
execute(...)method takes anExecutionContextinstance and aRequestobject. The object defines the environment in which the processing is occurring, including information about the JAASSubjectandLoginContext. TheRequestobject describes the requested operations on the content, with different concrete subclasses representing each type of activity. Examples of commands include (but not limited to) getting a node, moving a node, creating a node, changing a node, and deleting a node. And, if the repository source is able to participate in JTA/JTS distributed transactions, then the RepositoryConnection must implement thegetXaResource()method by returning a validjavax.transaction.xa.XAResourceobject that can be used by the transaction monitor.
As an example, consider that we want JBoss DNA to give us access through JCR to the schema information contained in a
relational databases. We first have to develop a connector that allows us to interact with relational databases using JDBC.
That connector would contain a JdbcRepositorySource Java class that implements RepositorySource,
and that has all of the various JavaBean properties for setting the name of the driver class, URL, username, password,
and other properties. (Or we might have a JavaBean property that defines the JNDI name where we can find a JDBC
DataSource instance pointing to our JDBC database.)
So with this very high-level summary, let's dive a little deeper and look at the repository example.
Recall that the example repository application consists of a client application that sets up a repository service and the repositories defined in a configuration repository, allowing the user to pick a repository and interactively navigate the selected repository. Several repositories are set up, including several in-memory repositories and one federated repository that dynamically federates the content from the other repositories.
The example is comprised of 2 classes and 1 interface, located in the src/main/java directory:
org/jboss/example/dna/repositories/ConsoleInput.java
/RepositoryClient.java
/UserInterface.java
RepositoryClient is the class that contains the main application. It uses an instance of the
UserInterface interface to methods that will be called at runtime to obtain information about the
files that are imported into the in-memory repositories and the JAAS CallbackHandler implementation
that will be used by JAAS to prompt the user for authentication information. Finally, the ConsoleInput
is an implementation of this that creates a text user interface, allowing the user to operate the client from the command-line.
We can easily create a graphical implementation of UserInterface at a later date, or we can also create a mock
implementation for testing purposes that simulates a user entering data. This allows us to check the behavior of the client
automatically using conventional JUnit test cases, as demonstrated by the code in the src/test/java directory:
org/jboss/example/dna/sequencers/RepositoryClientTest.java
/RepositoryClientUsingJcrTest.java
If we look at the RepositoryClient code, there are a handful of methods that encapsulate the various activities.
Note
Some of the code samples included in this book have had some of the error handling and comments removed so that the code is more readable and concise.
The main(String[] argv) method is of course the method that is executed when the application is run. This code
creates the JBoss DNA configuration by loading it from a file.
// Set up the JAAS provider (IDTrust) and a policy file (which defines the "dna-jcr" login config name)
IDTrustConfiguration idtrustConfig = new IDTrustConfiguration();
try {
idtrustConfig.config("security/jaas.conf.xml");
} catch (Exception ex) {
throw new IllegalStateException(ex);
}
// Now configure the repository client component ...
RepositoryClient client = new RepositoryClient();
for (String arg : args) {
arg = arg.trim();
if (arg.equals("--api=jcr")) client.setApi(Api.JCR);
if (arg.equals("--api=dna")) client.setApi(Api.DNA);
if (arg.equals("--jaas")) client.setJaasContextName(JAAS_LOGIN_CONTEXT_NAME);
if (arg.startsWith("--jaas=") && arg.length() > 7) client.setJaasContextName(arg.substring(7).trim());
}
// And have it use a ConsoleInput user interface ...
client.setUserInterface(new ConsoleInput(client, args));
The first block sets up the JAAS provider to be the IDTrust library and a policy file that defines the "dna-jcr" JAAS configuration.
The second block of code instantiates the RepositoryClient and passes in some options determined from the command-line.
It then sets the user interface (which then executes its behavior, which we'll see below).
The startRepositories() method builds the JcrEngine component from the configuration, starts the engine,
and obtains the JCR javax.jcr.Repository instance that the client will use. Note that the client has not yet
obtained a javax.jcr.Session instance, since this will be done each time the client needs to access content from
the repository. (This is actually a common practice according to the JCR specification, since Sessions are intended to be
very lightweight.)
public void startRepositories() throws IOException, SAXException {
if (engine != null) return; // already started
// Load the configuration from a file, as provided by the user interface ...
JcrConfiguration configuration = new JcrConfiguration();
configuration.loadFrom(userInterface.getRepositoryConfiguration());
// Load the node types for each JCR repository, via a CND file. These could have been defined
// in the configuration file, but this approach is easy and allows us to define the node types
// using the CND format in one or multiple files.
String locationOfCndFiles = userInterface.getLocationOfCndFiles();
configuration.repository("Aircraft").addNodeTypes(locationOfCndFiles + "/aircraft.cnd");
configuration.repository("Cars").addNodeTypes(locationOfCndFiles + "/cars.cnd");
configuration.repository("Vehicles").addNodeTypes(locationOfCndFiles + "/vehicles.cnd");
// Now create the JCR engine ...
engine = configuration.build();
engine.start();
// For this example, we're using a couple of in-memory repositories (including one for the configuration repository).
// Normally, these would exist already and would simply be accessed. But in this example, we're going to
// populate these repositories here by importing from files. First do the configuration repository ...
String location = this.userInterface.getLocationOfRepositoryFiles();
// Now import the content for the two in-memory repositories ...
Graph cars = engine.getGraph("Cars");
cars.importXmlFrom(location + "/cars.xml").into("/");
Graph aircraft = engine.getGraph("Aircraft");
aircraft.importXmlFrom(location + "/aircraft.xml").into("/");
}
This method does a number of different things. First, it checks to make sure the repositories are not already running; if so
the method just returns. Then, it creates a JBoss DNA JcrConfiguration instance and loads the configuration
from a file provided by the user interface. The method then loads the node types for each of the repositories; this could
have been done in the configuration, but it would have made the configuration file larger and more difficult to understand.
It then creates the JcrEngine from the configuration and starts it. Finally, it obtains the location of the
content files from the user interface, and imports them into the "Cars" and "Aircraft" repositories. Again, this is
done to keep the example simple.
The shutdown() method of the example then logs out and requests the JcrEngine instance shuts down and, since that may take
a few moments (if there are any ongoing operations or enqueued activities) awaits for it to complete the shutdown.
public void shutdown() throws InterruptedException, LoginException {
logout();
if (engine == null) return;
try {
// Tell the engine to shut down, and then wait up to 5 seconds for it to complete...
engine.shutdown();
engine.awaitTermination(5, TimeUnit.SECONDS);
} finally {
engine = null;
}
}
A few of the other methods in the RepositoryClient class deal with the JAAS LoginContext.
When needed, the client will authenticate the user (by asking the user interface for a callback handler that will be called
when the authentication information is needed). The resulting authenticated LoginContext is wrapped
by a custom javax.jcr.Credentials implementation. As long as the Credentials implementation
has a getLoginContext() method that returns a LoginContext object, JBoss DNA's repository
implementation will use that context to create the javax.jcr.Session. (Of course, the javax.jcr.SimpleCredentials
can also be used to create a Session, and JBoss DNA will then attempt to use JAAS to authenticate the user given by the credentials.)
The getNodeInfo(...) method of the example is what is called when the properties and children of a particular node
are requested by the user interface. (In the console user interface, this happens when the user navigates the graph structure.)
There are really two different behaviors to this method, depending upon whether the JCR API is to be used or whether
the JBoss DNA Graph API is to be used. The portion that uses JCR is shown below:
JcrRepository jcrRepository = engine.getRepository(sourceName);
Session session = null;
if (loginContext != null) {
// Could also use SimpleCredentials(username,password) too
Credentials credentials = new JaasCredentials(loginContext);
session = jcrRepository.login(credentials);
} else {
session = jcrRepository.login();
}
try {
// Make the path relative to the root by removing the leading slash(es) ...
pathToNode = pathToNode.replaceAll("^/+", "");
// Get the node by path ...
Node root = session.getRootNode();
Node node = root;
if (pathToNode.length() != 0) {
if (!pathToNode.endsWith("]")) pathToNode = pathToNode + "[1]";
node = pathToNode.equals("") ? root : root.getNode(pathToNode);
}
// Now populate the properties and children ...
if (properties != null) {
for (PropertyIterator iter = node.getProperties(); iter.hasNext();) {
javax.jcr.Property property = iter.nextProperty();
Object[] values = null;
// Must call either 'getValue()' or 'getValues()' depending upon # of values
if (property.getDefinition().isMultiple()) {
Value[] jcrValues = property.getValues();
values = new String[jcrValues.length];
for (int i = 0; i < jcrValues.length; i++) {
values[i] = jcrValues[i].getString();
}
} else {
values = new Object[] {property.getValue().getString()};
}
properties.put(property.getName(), values);
}
}
if (children != null) {
// Figure out which children need same-name sibling indexes ...
Set<String> sameNameSiblings = new HashSet<String>();
for (NodeIterator iter = node.getNodes(); iter.hasNext();) {
javax.jcr.Node child = iter.nextNode();
if (child.getIndex() > 1) sameNameSiblings.add(child.getName());
}
for (NodeIterator iter = node.getNodes(); iter.hasNext();) {
javax.jcr.Node child = iter.nextNode();
String name = child.getName();
if (sameNameSiblings.contains(name)) name = name + "[" + child.getIndex() + "]";
children.add(name);
}
}
} catch (javax.jcr.ItemNotFoundException e) {
return false;
} catch (javax.jcr.PathNotFoundException e) {
return false;
} finally {
if (session != null) session.logout();
}
This code is literally just using the standard JCR API. First, it obtains a javax.jcr.Session instance
(using the available LoginContext), finds the desired javax.jcr.Node, copies the
properties and names of the children into collections supplied by the caller via method parameters, and
finally logs out of the session.
The JBoss DNA Graph API is actually an internal API used within the different components of JBoss DNA
(including the connector and sequencer frameworks), and provides low-level access to the exact same content.
Though we do not recommend using this API in your client applications, if you need to write a connector
or sequencer, you may need to know how to use the Graph API.
Here is the portion of the getNodeInfo(...) method that does the exact same operation as the JCR code
shown above:
// Use the DNA Graph API to read the properties and children of the node ...
ExecutionContext context = loginContext != null ? this.context.create(loginContext) : this.context;
Graph graph = engine.getGraph(context, sourceName);
graph.useWorkspace("default");
org.jboss.dna.graph.Node node = graph.getNodeAt(pathToNode);
if (properties != null) {
// Now copy the properties into the map provided as a method parameter ...
for (Property property : node.getProperties()) {
String name = property.getName().getString(context.getNamespaceRegistry());
properties.put(name, property.getValuesAsArray());
}
}
if (children != null) {
// And copy the names of the children into the list provided as a method parameter ...
for (Location child : node.getChildren()) {
String name = child.getPath().getLastSegment().getString(context.getNamespaceRegistry());
children.add(name);
}
}
Note that this code is significantly shorter than the equivalent code based upon the JCR API. This is in part because the Graph API doesn't have the notion of a stateful session. But some of it also is simply because the Graph API design requires less code to do the same kinds of operations.
None of the other methods in the RepositoryClient really do anything with JBoss DNA or JCR
per se. Instead, they really facilitate interaction with the user interface.
If we look at the ConsoleInput constructor, it starts the repository and a thread
for the user interface. At this point, the constructor returns, but the main application continues under the user interface thread.
When the user requests to quit, the user interface thread also shuts down the JCR repository.
public ConsoleInput( SequencerClient client ) {
try {
client.startRepositories();
System.out.println(getMenu());
Thread eventThread = new Thread(new Runnable() {
private boolean quit = false;
public void run() {
try {
while (!quit) {
// Display the prompt and process the requested operation ...
}
} finally {
try {
// Terminate ...
client.shutdown();
} catch (Exception err) {
System.out.println("Error shutting down repository: "
+ err.getLocalizedMessage());
err.printStackTrace(System.err);
}
}
}
});
eventThread.start();
} catch (Exception err) {
System.out.println("Error: " + err.getLocalizedMessage());
err.printStackTrace(System.err);
}
}
At this point, we've reviewed all of the interesting code in the example application related to JBoss DNA. However, feel free to play with the application, trying different things.
This chapter walked through running the repository example and looked at the example code. This example allowed you to walk through multiple repositories, including one whose content was federated from multiple other repositories. This was a very simplistic example that only took a few minutes to run.
In the next chapter we'll wrap up by summarizing what we've learned about JBoss DNA and provide information about where you can find out more about JBoss DNA.
This document provided a very high-level overview of JBoss DNA, introducing you to what would be required to use JBoss DNA
in your own application. We saw two simple examples that showed two different aspects of JBoss DNA: the sequencing system
and the connector system. Each of these examples showed how to create a JBoss DNA configuration, how to start up
the JBoss DNA engine, and how to then access a javax.jcr.Repository instance, and after that your application
just uses the standard JCR API.
For a more in-depth description of JBoss DNA and the internal workings, see our Reference Guide. This also describes how to write your own custom sequencers or connectors. If you have any questions or comments, please feel free to contact JBoss DNA's user mailing list or use the user forums or (preferably) the mailing lists. If you find a bug or have a suggestion, please create a new issue in the project's JIRA issue management system . If there's something in particular you're interested in, talk with the community - there may be others interested in the same thing.
What's next for JBoss DNA? Well, the sequencing system is just the beginning. With this release, the sequencing system is stable enough so that more sequencers can be developed and used within your own applications. We've also established the foundation for JBoss DNA repositories, including a number of connectors. We'll continue to expand our library of sequencers and connectors, as well as expand our support of JCR. Other components on our roadmap include a web user interface, a REST-ful server, and a view system that allows domain-specific views of information in the repository. These components are farther out on our roadmap, and at this time have not been targeted to a particular release.